Ellie Zolfagharifard
DNA footprints left in the wake of ancient warriors can unravel long-held secrets about a country’s past. Now, for the first time, scientists have mapped the DNA of 95 different populations to see if they can paint a clearer picture of major historical events.
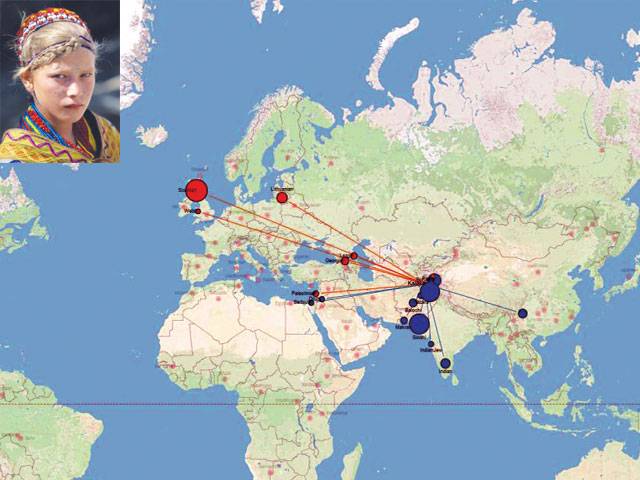
This incredible interactive map looks at the mixing of genes in major populations in Europe, Africa, Asia and South America spanning over the last four millennia.
The map reveals the genetic impact of European colonialism, the Arab slave trade, the Mongol Empire and European traders near the Silk Road mixing with people in China. ‘We made some very surprising discoveries,’ Dr Garrett Hellenthal of the University College London Genetics Institute told MailOnline. ‘For instance, the Kalash group is an isolated population in Pakistan. They are believed to be descendants of Alexander the Great’s invading army, and our map doesn’t dispute that fact.’
Many of the map’s genetic observations match historical events, and provide evidence of previously unrecorded genetic mixing. For example, the DNA of the Tu people in modern China suggests that in around 1200CE, Europeans similar to modern Greeks mixed with an otherwise Chinese-like population. Plausibly, said the researchers, the source of this European-like DNA might be merchants travelling the nearby Silk Road.
The powerful technique, dubbed ‘Globetrotter’, also provides insight into past events such as the genetic legacy of the Mongol Empire. Historical records suggest that the Hazara people of Pakistan are partially descended from Mongol warriors, and this study found clear evidence of Mongol DNA entering the population during the period of the Mongol Empire.
Six other populations, from as far west as Turkey, showed similar evidence of genetic mixing with Mongols around the same time. ‘What amazes me most is simply how well our technique works,’ said Dr Hellenthal. ‘Although individual mutations carry only weak signals about where a person is from, by adding information across the whole genome we can reconstruct these mixing events.
‘Sometimes individuals sampled from nearby regions can have surprisingly different sources of mixing.’ ‘For example, we identify distinct events happening at different times among groups sampled within Pakistan, with some inheriting DNA from sub-Saharan Africa, perhaps related to the Arab Slave Trade, others from East Asia, and yet another from ancient Europe. ‘Nearly all our populations show mixing events, so they are very common throughout recent history and often involve people migrating over large distances.’
The team at UCL and Oxford University used genome data for all 1490 individuals to identify ‘chunks’ of DNA that were shared between individuals from different populations. They were able to pinpoint time frame when genetic mixing took place using genetic recombination – the biological process in which two DNA molecules exchange genetic information.
Populations sharing more ancestry share more chunks, and individual chunks give clues about the underlying ancestry along chromosomes. ‘Each population has a particular genetic “palette”’, said Dr Daniel Falush of the Max Planck Institute for Evolutionary Anthropology in Leipzig, co-senior author of the study. ‘If you were to paint the genomes of people in modern-day Maya, for example, you would use a mixed palette with colours from Spanish-like, West African and Native American DNA.’
This mix dates back to around 1670CE, consistent with historical accounts describing Spanish and West African people entering the Americas around that time.
Though we can’t directly sample DNA from the groups that mixed in the past, we can capture much of the DNA of these original groups as persisting, within a mixed palette of modern-day groups,’ said Dr Falush. ‘This is a very exciting development.’
As well as providing fresh insights into historical events, the new research might have implications for how DNA impacts health and disease in different populations. ‘Understanding well the genetic similarities and differences between human populations is key for public health,’ said Dr Simon Myers.
Some populations are more at risk of certain diseases than others, and how well certain drugs work is also known to vary significantly. Rare genetic mutations are particularly likely to show strong differences between populations, and understanding their role in human health is an area of intense current research efforts.
The researchers hope that in the future they can include even more detailed sequencing, to spot these rare mutations and better understand their global spread. ‘This really is a fascinating insight,’ said Dr Hellenthal. ‘Imagine just how much more can be found out with even more detailed sequencing.’ –Daily Mail
Tuesday, April 16, 2024
From Mongol warriors to Silk Road traders
Caption: From Mongol warriors to Silk Road traders
IHC dismisses Bushra Bibi's plea seeking her shifting to Adiala Jail from Bani Gala
4:25 PM | April 16, 2024
Punjab CM visits Tehsil Headquarter Hospital Murree
3:25 PM | April 16, 2024
High-level Saudi delegation in Islamabad to hold meetings with Pakistani leadership
2:07 PM | April 16, 2024
Saudi foreign minister meets PM Shehbaz Sharif
1:17 PM | April 16, 2024
Decision to retaliate against Iran attacks rests with Israel, says Pentagon
1:05 PM | April 16, 2024
Political Reconciliation
April 16, 2024
Pricing Pressures
April 16, 2024
Western Hypocrisy
April 16, 2024
Policing Reforms
April 15, 2024
Storm Safety
April 15, 2024
Democratic harmony
April 16, 2024
Digital dilemma
April 16, 2024
Classroom crisis
April 16, 2024
Bridging gaps
April 16, 2024
Suicide awareness
April 15, 2024
ePaper - Nawaiwaqt
Advertisement
Nawaiwaqt Group | Copyright © 2024





